For a list of our publications, please visit the Cite Us page
jViz.RNA 4.0 - Quick Start
First, please download the files to be used in this guide by right clicking on them and selecting Save As from the drop down menu. Be sure to remember where you save them - your Desktop is a good location: short_RNA.ct long_RNA.ct pKnot_RNA.ct. These files are a collection of sample RNA molecules.
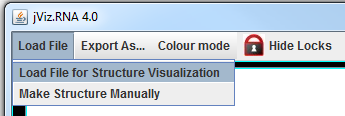
Download jViz.RNA 4.0 by visiting our Download page. To get started, load a structure or structures using the File menu. When the file chooser dialog pops up, select the appropriate .ct file to load. You can load multiple files one by one.
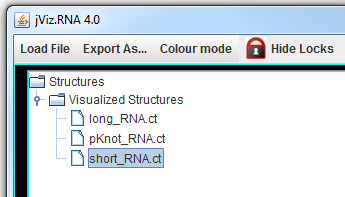
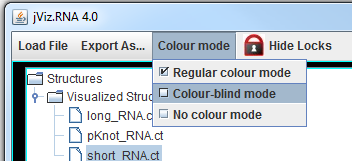
The left side of the screen contains the structure manager at the top, which allows you to switch between the structures.
jViz.RNA 4.0 also allows you to switch between colour schemes from the Colour Mode menu at the top. You can use these colour adjustments before exporting the structure as an image file.
jViz.RNA 2.0 - Quick Start
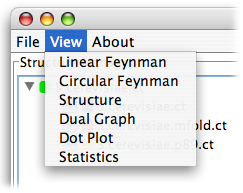
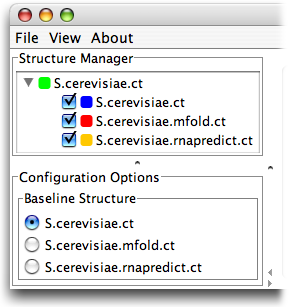
First, please download the files to be used in this guide by right clicking on them and selecting Save As from the drop down menu. Be sure to remember where you save them - your Desktop is a good location: S.cerevisiae.ct S.cerevisiae.rnapredict.ct S.cerevisiae.mfold.ct. These files are, in order, the native fold of Saccharomyces cerevisiae, a prediction from rnapredict, and a prediction from mfold.
Now start the program by clicking the following link to jViz.Rna 2.0. To get started, load a structure or structures using the File menu. When the file chooser dialog pops up, you can select multiple structures by holding the Shift or Ctrl keys to select a block of files or add or remove individual files to the current selection. jViz.Rna can load files with .ct, .bpseq, .dbf, .dbn, or .dbn.txt extensions.